Introduction:Pts who receive covalent Bruton tyrosine kinase inhibitors (cBTKis) for CLL can develop drug resistance, leading to disease progression. Often, cBTKi resistance results from the emergence of subclones with mutations at the cBTKi binding site (C481). Less frequent, non-C481 mutations, including gatekeeper residue T474 and kinase-impaired L528 mutations, have been reported in pts with progression on cBTKis. Most characterizations of cBTKi resistance mutations are derived from small or retrospective pt populations. To gain further insight into the genetic mechanisms of cBTKi resistance in a randomized population of pts with CLL, we performed next-generation sequencing (NGS) on samples from pts who had progression on zanubrutinib (zanu) or ibrutinib (ibr) in the phase 3 ALPINE study (NCT03734016; Brown et al. NEJM. 2023).
Methods:Progressive disease (PD) in ALPINE pts was assessed by independent review committee (IRC; n=139; zanu, n=54; ibr, n=85) and/or by investigator (INV; n=132; zanu, n=54; ibr, n=78) using Hallek et al ( Blood. 2008) criteria. Peripheral blood samples were collected at or after PD from 57 pts (zanu, n=26; ibr, n=31); 94.7% (54/57) had paired baseline samples. In 57 pts with PD, the median study follow-up was 25.7 mo; the median number of prior lines of therapy was 1 (range: zanu, 1-3; ibr, 1-7). Four pts (zanu, n=2; ibr, n=2) had Richter transformation (RT) at PD.
A high-sensitivity NGS panel (PredicineHEME) with full exon coverage of 106 genes, including BTK, PLCG2 and 27 putative CLL driver genes (Knisbacher et al. Nat Gen. 2022), was used for mutation analysis. All acquired mutations in BTK and PLCG2were reported at a variant allele frequency (VAF) of ≥0.25%. For all other genes, pathogenic mutations (assessed using VarSome) with a VAF of ≥1% are reported. Del(17p), del(11q), trisomy 12, del(13q), IGHV mutation status, and complex karyotype (CKT) status were assessed in baseline samples.
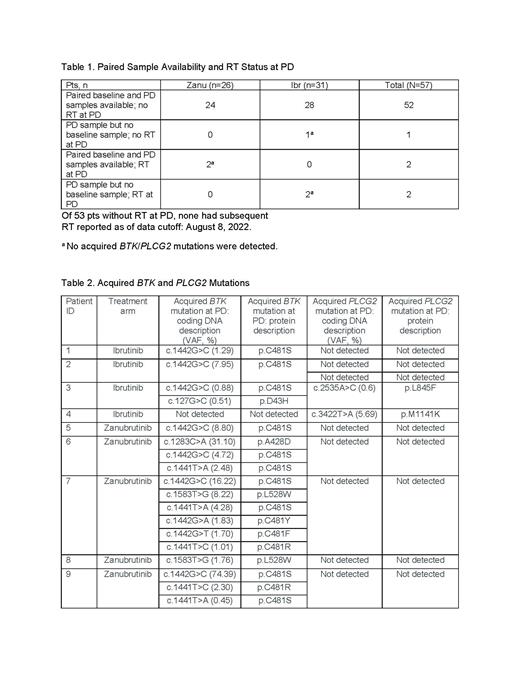
Results: No BTK mutations were detected at baseline, but 2 pts had PLCG2 mutations (zanu, R589C; ibr, E914STOP). To determine rates of BTK/PLCG2 or CLL driver gene mutations, only pts with paired baseline and PD samples and without RT at PD (n=52; zanu, n=24; ibr, n=28) were included (Table 1).
Nine pts acquired BTK/ PLCG2 mutations: 8 (zanu, n=5 [20.8%]; ibr, n=3 [10.7%]) had BTK mutations, and 2 (both in ibr; 7.1%) had PLCG2 mutations; 1 pt had both BTK and PLCG2 mutations. Of 18 single nucleotide variants (SNV) in BTK, 77.8% (n=14; zanu, n=11; ibr, n=3) were at C481. Non-C481 mutations were detected in 12.5% of pts (3/24) with progression on zanu (Table 2). All 9 pts with acquired BTK/ PLCG2 mutations had PD assessed by IRC and INV during treatment, except 1 ibr pt who had PD assessed by INV after treatment.
Median treatment duration was shorter in pts with PD without BTK mutations (zanu: n=19; 16.8 mo; range, 4.4-33.3 mo; ibr: n=25; 15.9 mo; range, 5.9-29.4 mo) than in pts with PD with acquired BTK mutations (zanu: n=5; 29.7 mo; range, 18.4-34.6 mo; ibr: n=3; 30.8 mo; range, 11.8-34.5 mo).
At baseline, SNV or indels in 18/27 driver genes were observed in 48/52 pts. Most frequent: NOTCH1 (n=21), TP53 (n=19), and ATM (n=8). Copy number aberrations (CNA) in 9/27 driver genes were observed in 23 pts. Most frequent: CCND2 (n=10, amplification [amp]), ATM (n=8, deletion [del]), TP53 (n=6, del), and KMT2D (n=6, amp). At PD, acquired SNV were observed in 3 pts ( SETD2, ASXL1, and SF3B1 n=1 each; all ibr). Acquired CNA in driver genes were observed in 10 pts ( KRAS amp: zanu, n=3; NRAS amp: ibr, n=2; CDKN1B amp: zanu, n=2, ibr, n=1; BIRC3 del: ibr, n=2). Acquisition of driver gene mutations was not associated with del(17p), IGHV mutation, or CKT status.
Conclusion: Of the 52 pts, most (82.6%) did not have acquired BTK or PLCG2 mutations. Among the zanu pts, 3/24 (12.5%) developed non-C481 BTK mutations. This rate was lower than that reported by Woyach et al (ICML 2023); shorter follow-up and fewer prior therapies in the ALPINE study may explain this discrepancy. Additionally, these data suggest that BTK and/or PLCG2 mutations are not the sole factors driving PD in this pt population; the presence or acquisition of mutations in driver genes with known negative prognostic value in CLL may also contribute to PD in pts without BTK or PLCG2 mutations. Given the low incidence of non-C481 mutations in ALPINE pts with PD, pts with CLL who were treated with cBTKis are likely to remain sensitive to other BTK-targeting therapies.
Disclosures
Brown:Gilead: Research Funding; Alloplex Biotherapeutics: Consultancy; Genentech/Roche: Consultancy; BeiGene: Consultancy, Research Funding; Abbvie: Consultancy; Acerta/AstraZeneca: Consultancy; Pharmacyclics: Consultancy; Pfizer: Consultancy; Numab Therapeutics: Consultancy; MEI Pharma: Research Funding; Merck: Consultancy; Loxo/Lilly: Consultancy, Research Funding; Kite: Consultancy; iOnctura: Consultancy, Research Funding; Hutchmed: Consultancy; Grifols Worldwide Operations: Consultancy; SecuraBio: Research Funding; TG Therapeutics: Research Funding. Li:BeiGene: Current Employment, Current equity holder in publicly-traded company. Eichhorst:F. Hoffmann-La Roche Ltd: Honoraria, Research Funding, Speakers Bureau; MSD: Consultancy, Honoraria, Speakers Bureau; Lilly: Consultancy, Speakers Bureau; Janssen: Consultancy, Research Funding, Speakers Bureau; Gilead: Consultancy, Research Funding; BeiGene: Consultancy, Honoraria, Research Funding, Speakers Bureau; AstraZeneca: Consultancy, Honoraria, Research Funding, Speakers Bureau; Abbvie: Consultancy, Honoraria, Research Funding, Speakers Bureau. Lamanna:Adaptive Biotechnologies: Consultancy; Eli Lilly/Loxo: Research Funding; Pharmacyclics: Consultancy; MingSight: Research Funding; Oncternal: Research Funding; Octapharma: Research Funding; Janssen: Consultancy; AstraZeneca: Consultancy, Research Funding; Genentech: Consultancy, Research Funding; Abbvie: Consultancy, Research Funding; BeiGene: Consultancy, Research Funding; TG Therapeutics: Research Funding. O'Brien:Astrazeneca: Consultancy; Beigene: Consultancy, Research Funding; Lilly: Consultancy, Research Funding; Janssen: Consultancy; Johnson & Johnson: Consultancy; Pfizer: Consultancy, Research Funding; Pharmacyclics: Consultancy, Research Funding; Regeneron: Research Funding; Abbvie: Consultancy. Tam:Janssen: Honoraria, Research Funding; AbbVie: Honoraria, Research Funding; BeiGene: Honoraria, Research Funding; LOXO: Honoraria; Novartis: Honoraria; Roche: Honoraria. Qiu:BeiGene, Janssen, Roche, AstraZeneca, Takeda: Speakers Bureau; Institute of Hematology & Blood Diseases Hospital, Chinese Academy of Medical Sciences: Current Employment. Ramakrishnan:BeiGene: Current equity holder in publicly-traded company, Ended employment in the past 24 months. Huang:BeiGene, Ltd: Current Employment. Shi:BeiGene Co. Ltd.: Current Employment. Idoine:BeiGene: Current Employment, Current equity holder in publicly-traded company, Divested equity in a private or publicly-traded company in the past 24 months. Salmi:BeiGene: Current Employment, Current equity holder in publicly-traded company. Cohen:BeiGene: Current Employment, Current equity holder in publicly-traded company, Divested equity in a private or publicly-traded company in the past 24 months. Shadman:Genmab: Consultancy, Research Funding; Genentech: Consultancy, Research Funding; Vincerx: Research Funding; Mustang Bio: Consultancy, Research Funding; Janssen: Consultancy; Fate Therapeutics: Consultancy; ADC therapeutics: Consultancy; Bristol Myers Squibb: Consultancy, Research Funding; MorphoSys/Incyte: Consultancy, Research Funding; Eli Lilly: Consultancy; Kite, a Gilead Company: Consultancy; AstraZeneca: Consultancy, Research Funding; BeiGene: Consultancy, Research Funding; Pharmacyclics: Consultancy, Research Funding; AbbVie: Consultancy, Research Funding; MEI Pharma: Consultancy; Regeneron: Consultancy; TG Therapeutics: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal